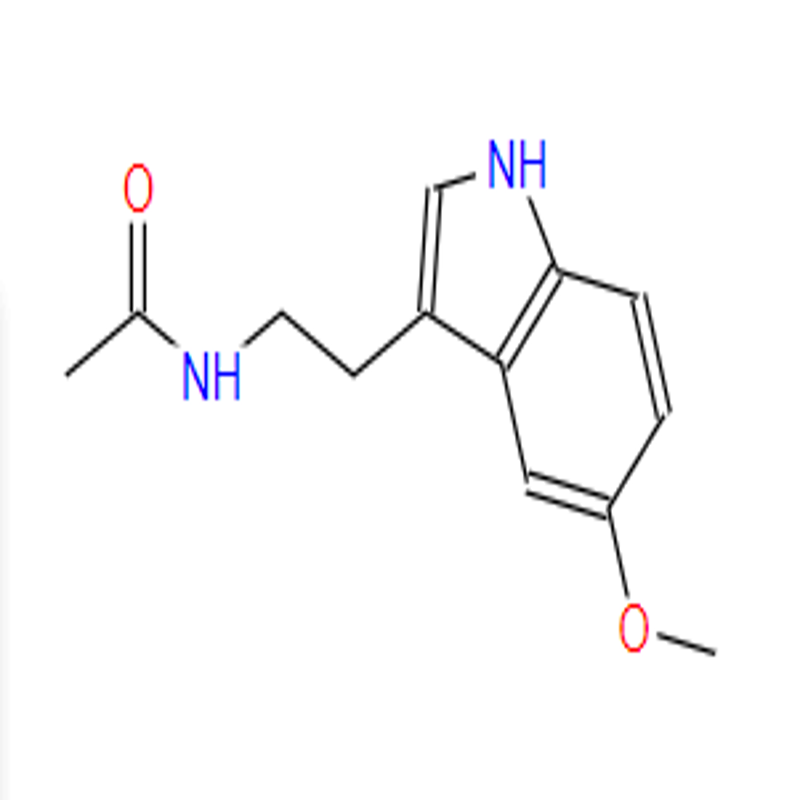
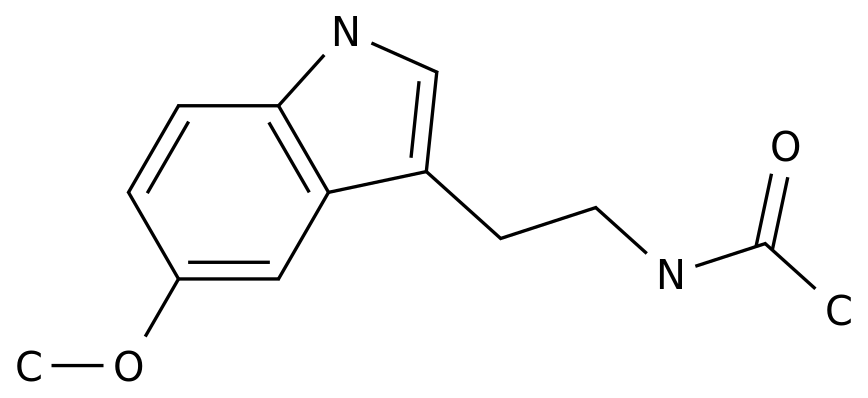
-
Categories
-
Pharmaceutical Intermediates
-
Active Pharmaceutical Ingredients
-
Food Additives
- Industrial Coatings
- Agrochemicals
- Dyes and Pigments
- Surfactant
- Flavors and Fragrances
- Chemical Reagents
- Catalyst and Auxiliary
- Natural Products
- Inorganic Chemistry
-
Organic Chemistry
-
Biochemical Engineering
- Analytical Chemistry
- Cosmetic Ingredient
-
Pharmaceutical Intermediates
Promotion
ECHEMI Mall
Wholesale
Weekly Price
Exhibition
News
-
Trade Service
Recently, Dr.
Chunmei Cai of Qinghai University School of Medicine was invited to publish a review paper titled: The Crosstalk between Viral RNA and DNA Sensing Mechanisms in the journal Cellular and Molecular Life Sciences.
The crossover between RNA and DNA recognition mechanisms in virus innate immunity and potential molecular mechanisms
.
Virus infection can cause many infectious and even fatal diseases, such as the current pandemic COVID-19, which poses a serious threat to humans
.
Therefore, it is more important to understand how the host's natural immune system recognizes viruses
.
The classic antiviral innate immunity is the production of type I interferon (IFN-I) and pro-inflammatory cytokines mediated by pattern recognition receptors (PRRs), thereby inducing a large number of ISGs to exert direct antiviral effects
.
Toll-like receptors that recognize viral nucleic acids are all located in the endosome
.
TLR3, TLR7 and TLR8 are RNA recognition receptors, and TLR9 is a DNA recognition receptor
.
RIG-I and MDA5 are responsible for recognizing viral RNA in the cytoplasm, while the detection of recognition receptors for cytoplasmic viral DNA is more complicated
.
cGAS is the main and indispensable receptor for detecting viral DNA in the cytoplasm, while IFI16 and DDX41 can only recognize the DNA of a few specific viruses
.
Although viral RNA and DNA recognition usually rely on different receptor and adaptor proteins, their downstream signal transduction is similar
.
It is worth noting that in antiviral innate immunity, there is a high degree of overlap between viral DNA and RNA recognition mechanisms, which play an important role in antiviral innate immunity
.
Figure 1.
DNA recognition receptor-mediated RNA virus detection.
Retroviruses are RNA viruses.
cGAS, DDX41, IFI16 and TLR9 can all recognize retrovirus-derived DNA and induce IFN-I production
.
Among them, cGAS is a major receptor that recognizes retroviral DNA, while DDX41, IFI16 and TLR9 can only recognize specific retrovirus-derived DNA
.
In addition, IFI16 can be used as a nucleic acid binding protein (RBP) and/or a regulatory protein, playing a key role in the strict regulation of the IFN/ISG signaling pathway
.
STING is a key adaptor protein for detecting cytoplasmic DNA.
It can directly transmit RIG-I-MAVS-mediated signals activated by RNA viruses to promote the production of IFN-I
.
Certain DNA viruses with AT-rich genomes can initiate RIG-I-MAVS-STING to induce the expression of IFN-I through the 5'-ppp dsRNA generated by transcription, and this process relies on RNA Pol III
.
In addition, STING can also inhibit the proliferation of RNA viruses by inhibiting the initiation of gene translation
.
It is worth noting that the early activation of STING limits Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) infection in vivo through IFN-dependent and independent mechanisms
.
On the contrary, SARS-CoV-2 inhibits the early activation of STING by encoding a large number of viral proteins, thereby evading the antiviral immunity mediated by STING
.
Figure 2.
Recognition of DNA virus by RNA recognition receptor RIG-I-mediated DNA virus detection is RNA Pol III dependent and independent
.
As mentioned above, RIG-I can recognize DNA viruses such as HSV-1 and EBV in an RNA Pol III-dependent manner
.
However, MDA5 and RIG-I detect KSHV-derived RNA in an RNA Pol III-independent manner
.
In addition, cytoplasmic RIG-I and MDA5, as well as endosome TLR3 and TLR8, can recognize RNA transcribed from DNA virus genome to limit viral infection, but whether DNA virus-derived RNA is RNA Pol III dependent, the specific mechanism remains Not sure
.
Viruses are still a major threat to human health, especially the SARS-CoV-2 strain that has been circulating in recent years
.
Although excellent progress has been made in understanding the innate immunity to viral infections mediated by multiple DNA/RNA recognition receptors, there are still many key unknowns regarding the overlap between DNA and RNA recognition mechanisms
.
The crossover between DNA and RNA recognition mechanisms enables the host to detect infection throughout the virus life cycle, thereby amplifying the innate antiviral response to RNA and DNA viruses
.
Interestingly, the virus has evolved a variety of mechanisms to suppress this overlap to evade the host's natural antiviral immunity
.
Therefore, advances in these fields will help develop viral vaccines or adjuvants and therapies that selectively target nucleic acid recognition
.
Journal introduction: Cellular and Molecular Life Sciences is a top journal in the field of biochemistry and molecular biology under Springer.
It was founded in 1997.
It mainly publishes the latest research and review articles in the fields of biology and biomedicine, including biochemistry and molecular biology, cell biology Science, biomedicine, neuroscience, pharmacology and immunology, with an annual volume of about 300 articles
.
Ranked 34th in the field of molecular biology
.
The latest impact factor in 2020 is 9.
261
.
First author: Dr.
Chunmei Cai (Bachelor of Sun Yat-Sen University, Doctor of Sun Yat-Sen University), Lecturer, Master's Supervisor, High Altitude Medicine Research Center, School of Medicine, Qinghai University
.
The main research direction is applied bioinformatics and systems biology, studying the correlation between glucose metabolism and natural immunity, the correlation between high altitude hypoxia adaptation and immune response, and exploring the potential of the innate immune system in the diagnosis and treatment of major diseases and high altitude endemic diseases.
Mechanism
.
Corresponding author: Professor Zheng Chunfu, School of Basic Medicine, Fujian Medical University
.
Paper link: https://pubmed.
ncbi.
nlm.
nih.
gov/34714359/ open for reprinting, welcome to forward to Moments and WeChat groups
Chunmei Cai of Qinghai University School of Medicine was invited to publish a review paper titled: The Crosstalk between Viral RNA and DNA Sensing Mechanisms in the journal Cellular and Molecular Life Sciences.
The crossover between RNA and DNA recognition mechanisms in virus innate immunity and potential molecular mechanisms
.
Virus infection can cause many infectious and even fatal diseases, such as the current pandemic COVID-19, which poses a serious threat to humans
.
Therefore, it is more important to understand how the host's natural immune system recognizes viruses
.
The classic antiviral innate immunity is the production of type I interferon (IFN-I) and pro-inflammatory cytokines mediated by pattern recognition receptors (PRRs), thereby inducing a large number of ISGs to exert direct antiviral effects
.
Toll-like receptors that recognize viral nucleic acids are all located in the endosome
.
TLR3, TLR7 and TLR8 are RNA recognition receptors, and TLR9 is a DNA recognition receptor
.
RIG-I and MDA5 are responsible for recognizing viral RNA in the cytoplasm, while the detection of recognition receptors for cytoplasmic viral DNA is more complicated
.
cGAS is the main and indispensable receptor for detecting viral DNA in the cytoplasm, while IFI16 and DDX41 can only recognize the DNA of a few specific viruses
.
Although viral RNA and DNA recognition usually rely on different receptor and adaptor proteins, their downstream signal transduction is similar
.
It is worth noting that in antiviral innate immunity, there is a high degree of overlap between viral DNA and RNA recognition mechanisms, which play an important role in antiviral innate immunity
.
Figure 1.
DNA recognition receptor-mediated RNA virus detection.
Retroviruses are RNA viruses.
cGAS, DDX41, IFI16 and TLR9 can all recognize retrovirus-derived DNA and induce IFN-I production
.
Among them, cGAS is a major receptor that recognizes retroviral DNA, while DDX41, IFI16 and TLR9 can only recognize specific retrovirus-derived DNA
.
In addition, IFI16 can be used as a nucleic acid binding protein (RBP) and/or a regulatory protein, playing a key role in the strict regulation of the IFN/ISG signaling pathway
.
STING is a key adaptor protein for detecting cytoplasmic DNA.
It can directly transmit RIG-I-MAVS-mediated signals activated by RNA viruses to promote the production of IFN-I
.
Certain DNA viruses with AT-rich genomes can initiate RIG-I-MAVS-STING to induce the expression of IFN-I through the 5'-ppp dsRNA generated by transcription, and this process relies on RNA Pol III
.
In addition, STING can also inhibit the proliferation of RNA viruses by inhibiting the initiation of gene translation
.
It is worth noting that the early activation of STING limits Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) infection in vivo through IFN-dependent and independent mechanisms
.
On the contrary, SARS-CoV-2 inhibits the early activation of STING by encoding a large number of viral proteins, thereby evading the antiviral immunity mediated by STING
.
Figure 2.
Recognition of DNA virus by RNA recognition receptor RIG-I-mediated DNA virus detection is RNA Pol III dependent and independent
.
As mentioned above, RIG-I can recognize DNA viruses such as HSV-1 and EBV in an RNA Pol III-dependent manner
.
However, MDA5 and RIG-I detect KSHV-derived RNA in an RNA Pol III-independent manner
.
In addition, cytoplasmic RIG-I and MDA5, as well as endosome TLR3 and TLR8, can recognize RNA transcribed from DNA virus genome to limit viral infection, but whether DNA virus-derived RNA is RNA Pol III dependent, the specific mechanism remains Not sure
.
Viruses are still a major threat to human health, especially the SARS-CoV-2 strain that has been circulating in recent years
.
Although excellent progress has been made in understanding the innate immunity to viral infections mediated by multiple DNA/RNA recognition receptors, there are still many key unknowns regarding the overlap between DNA and RNA recognition mechanisms
.
The crossover between DNA and RNA recognition mechanisms enables the host to detect infection throughout the virus life cycle, thereby amplifying the innate antiviral response to RNA and DNA viruses
.
Interestingly, the virus has evolved a variety of mechanisms to suppress this overlap to evade the host's natural antiviral immunity
.
Therefore, advances in these fields will help develop viral vaccines or adjuvants and therapies that selectively target nucleic acid recognition
.
Journal introduction: Cellular and Molecular Life Sciences is a top journal in the field of biochemistry and molecular biology under Springer.
It was founded in 1997.
It mainly publishes the latest research and review articles in the fields of biology and biomedicine, including biochemistry and molecular biology, cell biology Science, biomedicine, neuroscience, pharmacology and immunology, with an annual volume of about 300 articles
.
Ranked 34th in the field of molecular biology
.
The latest impact factor in 2020 is 9.
261
.
First author: Dr.
Chunmei Cai (Bachelor of Sun Yat-Sen University, Doctor of Sun Yat-Sen University), Lecturer, Master's Supervisor, High Altitude Medicine Research Center, School of Medicine, Qinghai University
.
The main research direction is applied bioinformatics and systems biology, studying the correlation between glucose metabolism and natural immunity, the correlation between high altitude hypoxia adaptation and immune response, and exploring the potential of the innate immune system in the diagnosis and treatment of major diseases and high altitude endemic diseases.
Mechanism
.
Corresponding author: Professor Zheng Chunfu, School of Basic Medicine, Fujian Medical University
.
Paper link: https://pubmed.
ncbi.
nlm.
nih.
gov/34714359/ open for reprinting, welcome to forward to Moments and WeChat groups