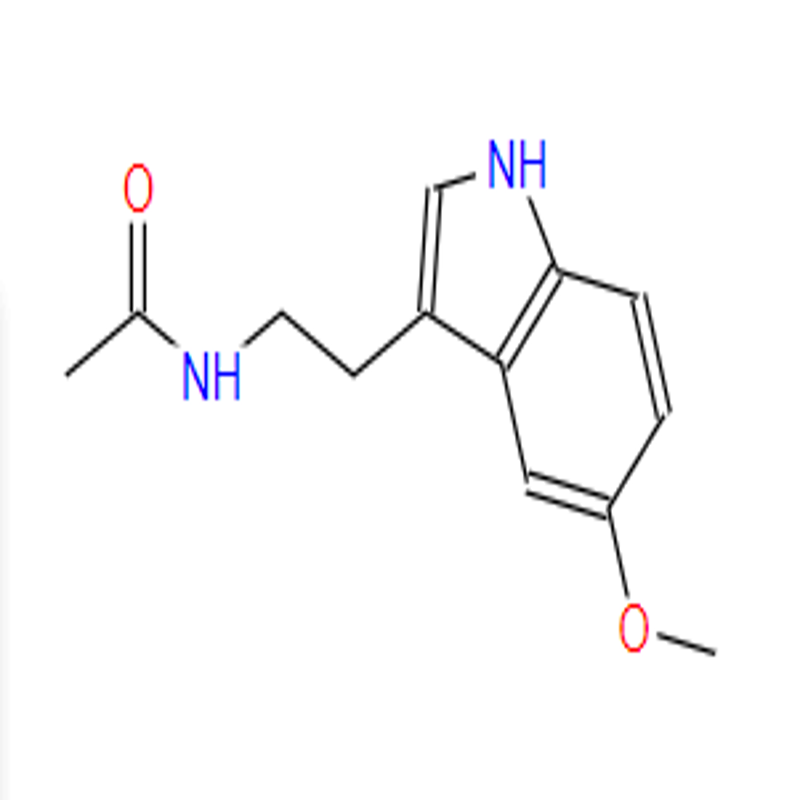
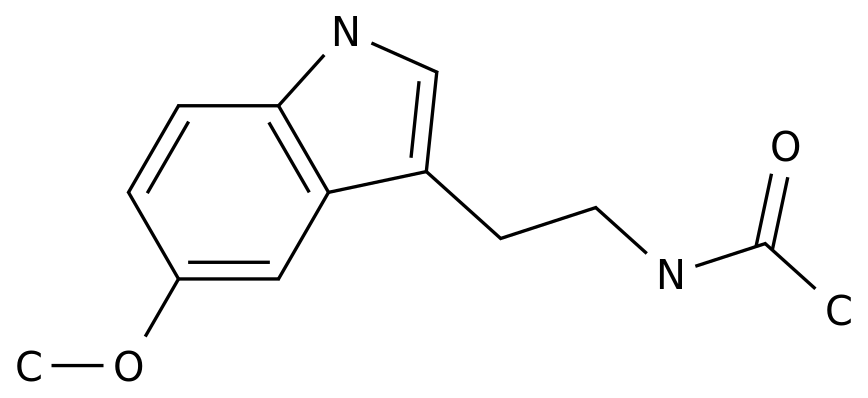
-
Categories
-
Pharmaceutical Intermediates
-
Active Pharmaceutical Ingredients
-
Food Additives
- Industrial Coatings
- Agrochemicals
- Dyes and Pigments
- Surfactant
- Flavors and Fragrances
- Chemical Reagents
- Catalyst and Auxiliary
- Natural Products
- Inorganic Chemistry
-
Organic Chemistry
-
Biochemical Engineering
- Analytical Chemistry
- Cosmetic Ingredient
-
Pharmaceutical Intermediates
Promotion
ECHEMI Mall
Wholesale
Weekly Price
Exhibition
News
-
Trade Service
The genetic difference between humans and chimpanzees is only 1 percent, but it is this 1 percent difference that distinguishes humans from
chimpanzees.
Recently, the team of Professor Zhu Hao of Southern Medical University in China published a research article
entitled "Uncovering the extensive trade-off between adaptive evolution and disease susceptibility" in the journal Cell Reports 。 The research team has developed a deep learning algorithm called DeepFavored, which can distinguish between favorable mutations and "free-rider" mutations by statistically testing existing genome-wide association study (GWAS) datasets
.
The article was published in Cell Reports
Because free-rider mutations are so similar to favorable mutations, it is difficult for any method to accurately distinguish between both as well as common mutations at the same time (Figure 1
).
Figure 1.
Subsequently, the research team used real-world GWAS data to detect genomic mutations in three populations: East Asia (CHB), Europe (CEU), and Africa (YRI), and compared the performance
of DeepFavored, iSAFE, and SWIF(r).
The researchers analyzed 1091 regions of the PopHumanScan gene in CEU, CHB, and YRI, which are good candidates for identifying favorable mutations
.
Figure 2.
After identifying favorable mutations in three human populations, the research team synthesized the correlation
between advantageous/free-rider mutations and GWAS sites.
To test the potential population-specificity and disease specificity of this gene mutation coordination, the research team analyzed the 454, 287, and 272 favorable mutations identified by DeepFavored in CEU, CHB, and YRI, and whether nearby free-rider mutations were rich in neurological, metabolic, and immune-related disease/trait sites
.
Figure 3: GWAS sites of different mutations
.
Environmental changes, pathogen evolution, and lifestyle changes drive adaptive evolution
of the genome.
Figure 4.
Resources:
1.
#secsectitle0080
2.
_msthash="320091" _msttexthash="67964">· END ·
: ,
。 Video Mini Program like, tap twice to cancel like in viewing, tap twice to cancel in viewing