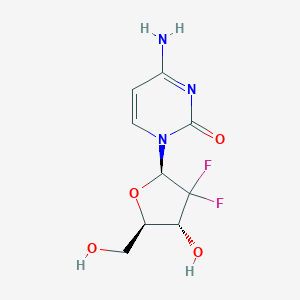
-
Categories
-
Pharmaceutical Intermediates
-
Active Pharmaceutical Ingredients
-
Food Additives
- Industrial Coatings
- Agrochemicals
- Dyes and Pigments
- Surfactant
- Flavors and Fragrances
- Chemical Reagents
- Catalyst and Auxiliary
- Natural Products
- Inorganic Chemistry
-
Organic Chemistry
-
Biochemical Engineering
- Analytical Chemistry
- Cosmetic Ingredient
-
Pharmaceutical Intermediates
Promotion
ECHEMI Mall
Wholesale
Weekly Price
Exhibition
News
-
Trade Service
Editor’s note iNature is China’s largest academic official account.
It is jointly created by the doctoral team of Tsinghua University, Harvard University, Chinese Academy of Sciences and other units.
The iNature Talent Official Account is now launched, focusing on talent recruitment, academic progress, scientific research information, and interested parties.
Long press or scan the QR code below to follow us.
Abnormal changes in iNature's epigenetic mechanisms (such as histone modifications) play an important role in cancer progression.
PRMT1, which triggers the asymmetric dimethylation of histidine H4 on arginine 3 (H4R3me2a), is upregulated in human colorectal cancer (CRC) and is essential for cell proliferation.
However, how this dysregulated modification may cause the malignant transformation of CRC is still poorly understood.
On April 14, 2021, Zhao Quan's team from the School of Life Sciences, Nanjing University published a research paper PRMT1-mediatedH4R3me2a recruits SMARCA4 to promote colorectal cancer progression by enhancing EGFR signaling in the internationally renowned journal Genome Medicine (IF=10.
68) in the field of genetics, revealing The histone H4R3me2a reader SMARCA4 in colorectal cancer cells maintains the expression of cancer-promoting genes and can be used as a new target for the treatment of colorectal cancer.
Doctoral students Yao Bing and Gui Tao from the School of Life Sciences of Nanjing University are the co-first authors of the paper, and Assistant Researcher Liu Ming and Professor Zhao Quan are the co-corresponding authors of the paper.
Colorectal carcinoma (CRC) is a malignant gastrointestinal tumor with poor prognosis caused by multiple pathogenic factors.
Morbidity and mortality are ranked third and fourth in malignant tumors, respectively.In addition to genetic factors, an important factor that drives the occurrence and development of CRC involves the accumulation of various epigenetic changes in intestinal epithelial cells.
Therefore, in-depth study of the molecular mechanism of epigenetic regulation of colorectal cancer has important guiding significance for the early diagnosis, clinical treatment and prognosis of colorectal cancer.
Protein arginine methyltransferase 1 (PRMT1) is a member of the arginine methyltransferase family and is the most abundant arginine methyltransferase in mammals.
PRMT1 mainly catalyzes the asymmetric double methylation (H4R3me2a) on the third arginine of histone H4, which is usually a marker of transcription activation, and is involved in gene transcription control, mRNA splicing, protein stability regulation, DNA damage signal transduction, and cells Fate is determined.
PRMT1 is also involved in the interaction of many transcription factors and gene promoters.
The overexpression and abnormal splicing of PRMT1 directly affect the occurrence of tumors, including breast cancer, lung cancer, bladder cancer and leukemia.
However, to date, the molecular recognition mechanism of PRMT1-mediated H4R3me2a is poorly understood.
In colorectal cancer, the pathophysiological function of PRMT1 is largely unknown, although PRMT1 has been identified as a marker of poor prognosis for patients with colorectal cancer.
In this study, the researchers first identified H4R3me2a by biochemical methods that specifically bind to the SMARCA4 protein (also known as BRG1, the catalytic subunit of the SWI/SNF chromatin remodeling complex, using hydrolyzed ATP to provide mediation of chromosome structural remodeling changes.
The required energy) can be read and identified.
Further studies have found that H4R3me2a can transcriptionally activate the EGFR signaling pathway after recruiting SMARCA4, thereby promoting the proliferation and migration of colorectal cancer cells; in vivo and in vitro experiments have shown that PRMT1 and SMARCA4 can act synergistically to promote the development of colorectal cancer.
On this basis, the researchers used EGFR cetuximab in combination with the PRMT1 inhibitor AMI-1 and found that the growth of colorectal cancer in vivo was more effectively inhibited.
This research provides new ideas and strategies for the clinically optimized treatment of colorectal cancer.
Article pattern diagram (picture from Genome Medicine) This research project is supported by the National Natural Science Foundation of China, the China Postdoctoral Science Foundation and the Central University Fund Project.
Reference message: https://genomemedicine.
biomedcentral.
com/articles/10.
1186/s13073-021-00871-5
It is jointly created by the doctoral team of Tsinghua University, Harvard University, Chinese Academy of Sciences and other units.
The iNature Talent Official Account is now launched, focusing on talent recruitment, academic progress, scientific research information, and interested parties.
Long press or scan the QR code below to follow us.
Abnormal changes in iNature's epigenetic mechanisms (such as histone modifications) play an important role in cancer progression.
PRMT1, which triggers the asymmetric dimethylation of histidine H4 on arginine 3 (H4R3me2a), is upregulated in human colorectal cancer (CRC) and is essential for cell proliferation.
However, how this dysregulated modification may cause the malignant transformation of CRC is still poorly understood.
On April 14, 2021, Zhao Quan's team from the School of Life Sciences, Nanjing University published a research paper PRMT1-mediatedH4R3me2a recruits SMARCA4 to promote colorectal cancer progression by enhancing EGFR signaling in the internationally renowned journal Genome Medicine (IF=10.
68) in the field of genetics, revealing The histone H4R3me2a reader SMARCA4 in colorectal cancer cells maintains the expression of cancer-promoting genes and can be used as a new target for the treatment of colorectal cancer.
Doctoral students Yao Bing and Gui Tao from the School of Life Sciences of Nanjing University are the co-first authors of the paper, and Assistant Researcher Liu Ming and Professor Zhao Quan are the co-corresponding authors of the paper.
Colorectal carcinoma (CRC) is a malignant gastrointestinal tumor with poor prognosis caused by multiple pathogenic factors.
Morbidity and mortality are ranked third and fourth in malignant tumors, respectively.In addition to genetic factors, an important factor that drives the occurrence and development of CRC involves the accumulation of various epigenetic changes in intestinal epithelial cells.
Therefore, in-depth study of the molecular mechanism of epigenetic regulation of colorectal cancer has important guiding significance for the early diagnosis, clinical treatment and prognosis of colorectal cancer.
Protein arginine methyltransferase 1 (PRMT1) is a member of the arginine methyltransferase family and is the most abundant arginine methyltransferase in mammals.
PRMT1 mainly catalyzes the asymmetric double methylation (H4R3me2a) on the third arginine of histone H4, which is usually a marker of transcription activation, and is involved in gene transcription control, mRNA splicing, protein stability regulation, DNA damage signal transduction, and cells Fate is determined.
PRMT1 is also involved in the interaction of many transcription factors and gene promoters.
The overexpression and abnormal splicing of PRMT1 directly affect the occurrence of tumors, including breast cancer, lung cancer, bladder cancer and leukemia.
However, to date, the molecular recognition mechanism of PRMT1-mediated H4R3me2a is poorly understood.
In colorectal cancer, the pathophysiological function of PRMT1 is largely unknown, although PRMT1 has been identified as a marker of poor prognosis for patients with colorectal cancer.
In this study, the researchers first identified H4R3me2a by biochemical methods that specifically bind to the SMARCA4 protein (also known as BRG1, the catalytic subunit of the SWI/SNF chromatin remodeling complex, using hydrolyzed ATP to provide mediation of chromosome structural remodeling changes.
The required energy) can be read and identified.
Further studies have found that H4R3me2a can transcriptionally activate the EGFR signaling pathway after recruiting SMARCA4, thereby promoting the proliferation and migration of colorectal cancer cells; in vivo and in vitro experiments have shown that PRMT1 and SMARCA4 can act synergistically to promote the development of colorectal cancer.
On this basis, the researchers used EGFR cetuximab in combination with the PRMT1 inhibitor AMI-1 and found that the growth of colorectal cancer in vivo was more effectively inhibited.
This research provides new ideas and strategies for the clinically optimized treatment of colorectal cancer.
Article pattern diagram (picture from Genome Medicine) This research project is supported by the National Natural Science Foundation of China, the China Postdoctoral Science Foundation and the Central University Fund Project.
Reference message: https://genomemedicine.
biomedcentral.
com/articles/10.
1186/s13073-021-00871-5