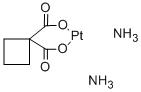
-
Categories
-
Pharmaceutical Intermediates
-
Active Pharmaceutical Ingredients
-
Food Additives
- Industrial Coatings
- Agrochemicals
- Dyes and Pigments
- Surfactant
- Flavors and Fragrances
- Chemical Reagents
- Catalyst and Auxiliary
- Natural Products
- Inorganic Chemistry
-
Organic Chemistry
-
Biochemical Engineering
- Analytical Chemistry
- Cosmetic Ingredient
-
Pharmaceutical Intermediates
Promotion
ECHEMI Mall
Wholesale
Weekly Price
Exhibition
News
-
Trade Service
Recently, the team of Professor Feng Chen from the Department of Biostatistics, School of Public Health, Nanjing Medical University, published the latest results of the team's lung cancer omics data mining in the sub-Journal NPJ Precision Oncology under Nature [1].
This multi-omics study based on genome, epigenome, transcriptome, and proteome reveals that TNS3 and SEPT7 are related to the prognosis of long-term (>10 years) smoking cessation lung cancer patients at different molecular levels.
Smoking is the main environmental exposure factor that affects the occurrence of lung cancer.
However, people who have quit smoking for a long time (eg, those who have quit smoking for more than 10 years) are still likely to develop lung cancer.
As we all know, genetic factors can affect the occurrence and development of diseases.
So, is the prognosis of lung cancer patients who quit smoking for a long time affected by specific genetic factors? In addition, are the genetic factors related to its prognosis different from those of non-smokers and smokers? The study mainly carried out statistical analysis from four angles.
The statistical analysis flow chart is shown in the figure below.
Statistical analysis flow chart (1) Two-stage genome-wide prognostic association study: Researchers cooperated with the International Lung Cancer Research Consortium (ILCCO) to carry out two-stage clinical prognostic data and GWAS data for 1,299 long-term smoking cessation non-small cell lung cancer (NSCLC) patients Statistical Analysis.
The discovery set contains 566 people and comes from the Harvard Lung Cancer Cohort (HLCS).
The validation set contains 733 people from the MD Anderson Lung Cancer Cohort (MDACC), Liverpool Lung Cancer Cohort (Liverpool), Asturias Lung Cancer Cohort (CAPUA), Carrotin and Retinol Lung Cancer Intervention Study (CARET), Toronto Lung Cancer queue.
The classical Cox proportional hazard model was used to evaluate the prognostic effect of SNP.
The correlation results were corrected by multiple tests and verified by independent populations to control false positives.
In the end, the study identified two SNPs related to the prognosis of NSCLC patients with long-term smoking cessation: rs34211819 (HR=0.
73, P=3.
90×10-9) located on the TNS3 gene in the 7p12.
3 region is better related to the prognosis of patients, and The rs1143149 (HR=1.
36, P=9.
75×10-9) located on the SEPT7 gene in the 7p14.
2 region is associated with a worse prognosis.
The prognostic correlation study of rs34211819 and rs1143149 with NSCLC (2) Gene-quit smoking interaction analysis: Stratified analysis with different smoking cessation time as a stratified factor shows that the prognostic effects of rs34211819 and rs1143149 are significantly heterogeneous at different smoking cessation times ( rs34211819: heterogeneity P=0.
042; rs1143149: heterogeneity P=0.
034).
The Cox proportional hazard model shows that two SNPs have a significant interaction with the duration of smoking cessation.
As the duration of smoking cessation changes, the effect of SNPs on the prognostic outcome (HR) has changed significantly.
With the increase of smoking cessation time, the protection of rs34211819 The effect gradually increased (interaction P=8.
0×10-4); on the contrary, the risk effect of rs1143149 gradually increased (interaction P=0.
003).
Gene-quit smoking duration interaction diagram (3) Multi-omics analysis: further, based on TCGA, CPTAC, HLCS and other databases, starting from the two gene levels of TNS3 and SEPT7, the QTL regulatory relationship of SNP and the gene expression and DNA The relationship between basicity, protein abundance and the prognosis of lung cancer.
Studies have found that high gene expression of TNS3 and SEPT7 are associated with poor prognosis of lung cancer, and their protein abundance in tumor tissues is significantly higher than that in normal tissues.
Two methylated CpG sites in the gene region are associated with survival outcomes.
It is also significantly related.
TNS3, SEPT7 multi-omics lung cancer prognosis analysis chart (4) GWAS genetic similarity comparison: identify prognostic-related SNPs in the subgroups of non-smokers, short-term smoking cessation, long-term smoking cessation, and smoking NSCLC.
Further compare the genetic similarity of signals at the level of SNP, gene, expression quantitative trait locus (eQTL), enhancer and pathway.
The results suggest that the genetic similarity between long-term quitters and non-smokers is higher than that of short-term quitters or current smokers.
From a genetic point of view, the prognosis of patients who quit smoking for a long time is more similar to the prognosis of non-smokers, which is precision medicine.
Provide theoretical basis.
Pathway enrichment analysis showed that the unique genes of NSCLC patients with long-term smoking cessation were significantly enriched in immune-related pathways, such as B cell receptor pathway and Th1/Th2 cell differentiation pathway.
In general, this study provides for the first time multiple levels of molecular evidence related to the prognosis of NSCLC patients with long-term smoking cessation, and reveals the specificity of their genetic structure, providing important clues for drug development.
Dr.
Shen Sipeng and Associate Professor Wei Yongyue from the Department of Biostatistics, School of Public Health, Nanjing Medical University are the co-first authors of the paper.
Associate Professor Zhang Ruyang, Professor Chen Feng and Professor David C.
Christiani from the Harvard School of Public Health are the co-corresponding authors of the paper.
In addition, this research was also supported by Professor Christopher I.
Amos from Baylor College of Medicine, Professor Geoffrey Liu from Princess Margaret Cancer Center, Canada, Professor Rayjean J.
Hung from University of Toronto, Canada, Professor John K.
Field from the University of Liverpool, UK, and Frederick Frederick, USA Professor Chu Chen from Jinsen Cancer Research Center, Professor Adonina Tardon from the University of Oviedo in Spain, Professor Yi Li from the University of Michigan in the United States, Professor Hongbing Shen, Professor Zhibin Hu, and Professor Yang Zhao from Nanjing Medical University.
References: [1] Responsible EditorBioTalker
This multi-omics study based on genome, epigenome, transcriptome, and proteome reveals that TNS3 and SEPT7 are related to the prognosis of long-term (>10 years) smoking cessation lung cancer patients at different molecular levels.
Smoking is the main environmental exposure factor that affects the occurrence of lung cancer.
However, people who have quit smoking for a long time (eg, those who have quit smoking for more than 10 years) are still likely to develop lung cancer.
As we all know, genetic factors can affect the occurrence and development of diseases.
So, is the prognosis of lung cancer patients who quit smoking for a long time affected by specific genetic factors? In addition, are the genetic factors related to its prognosis different from those of non-smokers and smokers? The study mainly carried out statistical analysis from four angles.
The statistical analysis flow chart is shown in the figure below.
Statistical analysis flow chart (1) Two-stage genome-wide prognostic association study: Researchers cooperated with the International Lung Cancer Research Consortium (ILCCO) to carry out two-stage clinical prognostic data and GWAS data for 1,299 long-term smoking cessation non-small cell lung cancer (NSCLC) patients Statistical Analysis.
The discovery set contains 566 people and comes from the Harvard Lung Cancer Cohort (HLCS).
The validation set contains 733 people from the MD Anderson Lung Cancer Cohort (MDACC), Liverpool Lung Cancer Cohort (Liverpool), Asturias Lung Cancer Cohort (CAPUA), Carrotin and Retinol Lung Cancer Intervention Study (CARET), Toronto Lung Cancer queue.
The classical Cox proportional hazard model was used to evaluate the prognostic effect of SNP.
The correlation results were corrected by multiple tests and verified by independent populations to control false positives.
In the end, the study identified two SNPs related to the prognosis of NSCLC patients with long-term smoking cessation: rs34211819 (HR=0.
73, P=3.
90×10-9) located on the TNS3 gene in the 7p12.
3 region is better related to the prognosis of patients, and The rs1143149 (HR=1.
36, P=9.
75×10-9) located on the SEPT7 gene in the 7p14.
2 region is associated with a worse prognosis.
The prognostic correlation study of rs34211819 and rs1143149 with NSCLC (2) Gene-quit smoking interaction analysis: Stratified analysis with different smoking cessation time as a stratified factor shows that the prognostic effects of rs34211819 and rs1143149 are significantly heterogeneous at different smoking cessation times ( rs34211819: heterogeneity P=0.
042; rs1143149: heterogeneity P=0.
034).
The Cox proportional hazard model shows that two SNPs have a significant interaction with the duration of smoking cessation.
As the duration of smoking cessation changes, the effect of SNPs on the prognostic outcome (HR) has changed significantly.
With the increase of smoking cessation time, the protection of rs34211819 The effect gradually increased (interaction P=8.
0×10-4); on the contrary, the risk effect of rs1143149 gradually increased (interaction P=0.
003).
Gene-quit smoking duration interaction diagram (3) Multi-omics analysis: further, based on TCGA, CPTAC, HLCS and other databases, starting from the two gene levels of TNS3 and SEPT7, the QTL regulatory relationship of SNP and the gene expression and DNA The relationship between basicity, protein abundance and the prognosis of lung cancer.
Studies have found that high gene expression of TNS3 and SEPT7 are associated with poor prognosis of lung cancer, and their protein abundance in tumor tissues is significantly higher than that in normal tissues.
Two methylated CpG sites in the gene region are associated with survival outcomes.
It is also significantly related.
TNS3, SEPT7 multi-omics lung cancer prognosis analysis chart (4) GWAS genetic similarity comparison: identify prognostic-related SNPs in the subgroups of non-smokers, short-term smoking cessation, long-term smoking cessation, and smoking NSCLC.
Further compare the genetic similarity of signals at the level of SNP, gene, expression quantitative trait locus (eQTL), enhancer and pathway.
The results suggest that the genetic similarity between long-term quitters and non-smokers is higher than that of short-term quitters or current smokers.
From a genetic point of view, the prognosis of patients who quit smoking for a long time is more similar to the prognosis of non-smokers, which is precision medicine.
Provide theoretical basis.
Pathway enrichment analysis showed that the unique genes of NSCLC patients with long-term smoking cessation were significantly enriched in immune-related pathways, such as B cell receptor pathway and Th1/Th2 cell differentiation pathway.
In general, this study provides for the first time multiple levels of molecular evidence related to the prognosis of NSCLC patients with long-term smoking cessation, and reveals the specificity of their genetic structure, providing important clues for drug development.
Dr.
Shen Sipeng and Associate Professor Wei Yongyue from the Department of Biostatistics, School of Public Health, Nanjing Medical University are the co-first authors of the paper.
Associate Professor Zhang Ruyang, Professor Chen Feng and Professor David C.
Christiani from the Harvard School of Public Health are the co-corresponding authors of the paper.
In addition, this research was also supported by Professor Christopher I.
Amos from Baylor College of Medicine, Professor Geoffrey Liu from Princess Margaret Cancer Center, Canada, Professor Rayjean J.
Hung from University of Toronto, Canada, Professor John K.
Field from the University of Liverpool, UK, and Frederick Frederick, USA Professor Chu Chen from Jinsen Cancer Research Center, Professor Adonina Tardon from the University of Oviedo in Spain, Professor Yi Li from the University of Michigan in the United States, Professor Hongbing Shen, Professor Zhibin Hu, and Professor Yang Zhao from Nanjing Medical University.
References: [1] Responsible EditorBioTalker