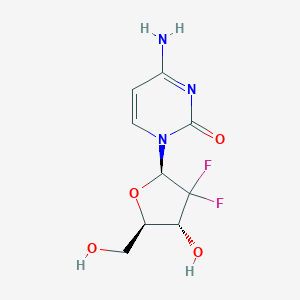
-
Categories
-
Pharmaceutical Intermediates
-
Active Pharmaceutical Ingredients
-
Food Additives
- Industrial Coatings
- Agrochemicals
- Dyes and Pigments
- Surfactant
- Flavors and Fragrances
- Chemical Reagents
- Catalyst and Auxiliary
- Natural Products
- Inorganic Chemistry
-
Organic Chemistry
-
Biochemical Engineering
- Analytical Chemistry
- Cosmetic Ingredient
-
Pharmaceutical Intermediates
Promotion
ECHEMI Mall
Wholesale
Weekly Price
Exhibition
News
-
Trade Service
There are currently more than 100 RNA modifications known, of which RNA methylation modifications account for more than 60%, and m6A (N6-methyladenine), as the most common modification on RNA in higher organisms, has only increased in popularity in recent years.
Minus
.
Our public account has also pushed a lot of articles in this series for you.
Today, I will continue to look at two articles published in the same issue of Molecular Therapy-Nucleic Acids (IF=7.
032) in April.
Both articles discuss m6A modification.
The role of lncRNAs in tumors
.
The m6A modification mainly occurs on the adenine in the RRACH sequence, and its function is determined by the encoder (methyltransferase), the reader (binding protein), and the decoder (demethylase) (Figure 1)
.
The encoder consists of METTL3, METTL14, WTAP, RBM15, ZC3H13, VIRMA, CBLL1 and the newly discovered METTL16
.
The m6A barcode reader contains binding proteins such as YTHDF1–3, YTHDC1–2, IGF2BPs, HNRNPs, eIF3, FMRP, Prrc2a, METTL3 and LRPPRC, which can specifically recognize m6A methyltransferase and affect methylated mRNAs, different Binding proteins have different functions
.
Decoder is responsible for regulating the demethylation process of m6A, including FTO, ALKBH5, ALKBH9 and ALKBH10B proteins
.
Figure 1.
Composition of m6A The author of the first article Yufei Lan et al.
summarized the regulatory mechanism of m6A modification on lncRNA function (Figure 2)
.
On the one hand, m6A modification acts on the RNA-DNA triple helix structure to regulate the relationship between lncRNA and specific DNA sites; on the other hand, m6A modification provides binding sites for barcode readers or regulates the structure of local RNA, thereby inducing RBPs The combination of lncRNA regulates the function of lncRNA
.
Figure 2.
The regulatory mechanism of m6A on lncRNAs.
Multiple studies have shown that both m6A and lncRNA play a certain role in the occurrence and development of tumors (Figure 3).
m6A modified lncRNA can induce pancreatic cancer, ovarian cancer, liver cancer and other tumor cells.
Proliferation, migration and apoptosis
.
For example, Wu et al.
found that m6A-induced lncRNA-RP11 can trigger the metastasis and proliferation of colorectal cancer cells by up-regulating Zeb1; IGF2BP2 can act on the cell proliferation and tumorigenesis of pancreatic cancer by regulating the expression of lncRNA DANCR and DANCR.
The study of He et al.
found that ALKBH5 can inhibit pancreatic cancer by demethylating lncRNA KCNK15-AS1 and regulating its expression, providing a potential target for the treatment of pancreatic cancer
.
Figure 3.
m6A-modified lncRNAs in tumors.
Feng Xu, the author of the second article, used the expression profiles of lncRNAs and m6A genes in TCGA and data from patients with LUAD (lung adenocarcinoma) to identify m6A-related lncRNAs as the prognosis of LUAD And potential markers of immune response (Figure 4)
.
The author first used Pearson correlation analysis to screen out lncRNAs related to 21 m6A genes; then, the LUAD data set was randomly divided into a training set and a validation set, combined with clinical information, and further screened lncRNAs related to OS through univariate Cox regression analysis , And used LASSO Cox regression analysis to identify 24 m6A-related lncRNAs that are significantly related to OS; next, a multivariate Cox regression analysis was used to establish a risk model based on 12-m6A-related lncRNAs, and based on the median risk The scores are low-risk group and high-risk group, and Kaplan-Meier is used to analyze the difference in OS between the two groups
.
In addition, drug susceptibility data was used to discover drug candidates targeting these lncRNAs
.
Figure 4.
Flow chart.
Setting the threshold |Pearson R|> 0.
3 & p <0.
001, the author identified 1,149 m6A-related lncRNAs, and the co-expression network is shown in Figure 5
.
Figure 5.
Identifying m6A-related lncRNAs Using univariate Cox regression analysis, 38 m6A-related lncRNAs that are significantly related to OS were screened in the TCGA data set (Figure 6A)
.
LASSO-penalized Cox analysis screened out 24 m6A-related lncRNAs (Figure 6B-C) for subsequent multivariate Cox ratio risk regression analysis, and finally used to construct a risk model to assess the prognostic risk of LUAD patients.
The 12 lncRNAs are shown in Figure 6D Shown
.
Figure 6.
Risk model Figure 7A shows the distribution of risk levels between the low-risk and high-risk groups of the LUAD sample.
The survival status and survival time of the two groups of patients are shown in Figure 7B
.
The results of survival analysis showed that the survival period of the low-risk group was longer than that of the high-risk group (Figure 7D)
.
Figure 7.
The prognostic value of the risk model in the training set is to detect the prognostic performance of the risk model.
The study respectively depicts the risk level distribution, survival status and survival of the test set (Figure 8A-C) and the entire data set (Figure 8E-G) Time pattern, and the expression of m6A-related lncRNAs
.
The Kaplan-Meier survival analysis results of the test set and the entire data set are the same as the training set.
The OS of LUAD patients with higher risk scores is lower than that of patients with lower risk scores (Figure 8D-H)
.
Figure 8.
The prognostic value of the test concentration risk model.
After grouping the entire TCGA data set by gender, age, grade, stage and other clinical indicators, the study found that the OS of the low-risk group was still longer than that of the high-risk group (Figure 9)
.
Figure 9.
Kaplan-Meier survival curve.
Based on the entire gene expression profile, 21 m6A genes, 12 m6A-related lncRNAs, and risk models, the authors performed PCA to analyze the differences between the low-risk group and the high-risk group
.
It is found that in the results obtained based on the risk model, the high and low risk groups have different distributions (Figure 10D)
.
Figure 10.
Principal component analysis Based on the m6A-related lncRNA model obtained from 504 LUAD samples, the author further analyzed the enrichment level, activity and function of several immune cells and pathways in LUAD, and found the immune indicators of the low-risk group and the high-risk group The expression difference was significant (Figure 11A)
.
In addition, through the correlation analysis between the m6A-related lncRNA model and immunotherapy markers, the study found that the high-risk group responded better to immunotherapy (Figure 11C), indicating that the model can be used as a predictor of tumor immune dysfunction and rejection Indicators
.
Next, the author used the R package maftools to analyze the mutation data.
Figure 11D and Figure 11E respectively show the top 20 driver genes with the highest mutation frequency in the high and low risk groups.
.
Then, the TMB score was calculated based on the TGCA somatic mutation data, and the results showed that the classification index has a high correlation with TMB (Figure 11F)
.
The author also used IC50 data in the GDSC database to screen out 78 compounds based on thepRRophetic algorithm as potential therapeutic drugs for LUAD targeting the lncRNA model
.
Figure 11.
Using the model to evaluate immunotherapy response.
The results of univariate and multivariate Cox regression analysis showed that the risk model was independent of other clinical indicators and confirmed its reliability (Figure 12)
.
Figure 12.
Evaluation of the prognostic risk model The authors used a nomogram containing risk levels and clinical risk characteristics to predict the incidence of OS at 1, 2 and 3 years
.
Compared with clinical indicators, the predictive model showed significant predictive ability (Figure 13A)
.
From the correlation diagram in Figure 13B-D, it can be found that the observation rates of 1-year, 2-year, and 3-year OS are consistent with the predicted rates
.
Figure 13.
Construction and evaluation of prognostic nomograms With the development of MeRIP-seq and m6A sequencing technologies, more m6A-modified lncRNAs will be discovered, and their functional significance needs to be explored
.
In addition, research on m6A-related proteins and their inhibitors will also provide new opportunities for early diagnosis, treatment and even prognosis of cancer
.
Therefore, for m6A, there is still a lot of articles to do.
I hope that the articles shared today can bring a bit of inspiration to interested friends
.
Have a nice day! Reference articles: 1.
The role of M6A modification in the regulation of tumor-related lncRNAs (PMID: 33996258) 2.
m6A-related lncRNAs are potential biomarkers for predicting prognoses and immune responses in patients with LUAD (PMID: 33996259) If you have questions about life information, please contact: 18501230653 Dragon Boat Festival Ankang Transcriptome | Methylation | Resequencing | Single Cell | m6A | Multiomics Cytoscape | limma | WGCNA | Water Bear Legend | Linux Electrophoresis | PCR | A Brief History of Sequencing | Karyotype | NIPT | Basic Experimental Genes | 2019-nCoV | Enrichment Analysis | Joint Analysis | Microenvironmental Plague Pursuit | Summary of Ideas | Scholars | Scientific Research | Retracted Papers | PhD Reading | Work
Minus
.
Our public account has also pushed a lot of articles in this series for you.
Today, I will continue to look at two articles published in the same issue of Molecular Therapy-Nucleic Acids (IF=7.
032) in April.
Both articles discuss m6A modification.
The role of lncRNAs in tumors
.
The m6A modification mainly occurs on the adenine in the RRACH sequence, and its function is determined by the encoder (methyltransferase), the reader (binding protein), and the decoder (demethylase) (Figure 1)
.
The encoder consists of METTL3, METTL14, WTAP, RBM15, ZC3H13, VIRMA, CBLL1 and the newly discovered METTL16
.
The m6A barcode reader contains binding proteins such as YTHDF1–3, YTHDC1–2, IGF2BPs, HNRNPs, eIF3, FMRP, Prrc2a, METTL3 and LRPPRC, which can specifically recognize m6A methyltransferase and affect methylated mRNAs, different Binding proteins have different functions
.
Decoder is responsible for regulating the demethylation process of m6A, including FTO, ALKBH5, ALKBH9 and ALKBH10B proteins
.
Figure 1.
Composition of m6A The author of the first article Yufei Lan et al.
summarized the regulatory mechanism of m6A modification on lncRNA function (Figure 2)
.
On the one hand, m6A modification acts on the RNA-DNA triple helix structure to regulate the relationship between lncRNA and specific DNA sites; on the other hand, m6A modification provides binding sites for barcode readers or regulates the structure of local RNA, thereby inducing RBPs The combination of lncRNA regulates the function of lncRNA
.
Figure 2.
The regulatory mechanism of m6A on lncRNAs.
Multiple studies have shown that both m6A and lncRNA play a certain role in the occurrence and development of tumors (Figure 3).
m6A modified lncRNA can induce pancreatic cancer, ovarian cancer, liver cancer and other tumor cells.
Proliferation, migration and apoptosis
.
For example, Wu et al.
found that m6A-induced lncRNA-RP11 can trigger the metastasis and proliferation of colorectal cancer cells by up-regulating Zeb1; IGF2BP2 can act on the cell proliferation and tumorigenesis of pancreatic cancer by regulating the expression of lncRNA DANCR and DANCR.
The study of He et al.
found that ALKBH5 can inhibit pancreatic cancer by demethylating lncRNA KCNK15-AS1 and regulating its expression, providing a potential target for the treatment of pancreatic cancer
.
Figure 3.
m6A-modified lncRNAs in tumors.
Feng Xu, the author of the second article, used the expression profiles of lncRNAs and m6A genes in TCGA and data from patients with LUAD (lung adenocarcinoma) to identify m6A-related lncRNAs as the prognosis of LUAD And potential markers of immune response (Figure 4)
.
The author first used Pearson correlation analysis to screen out lncRNAs related to 21 m6A genes; then, the LUAD data set was randomly divided into a training set and a validation set, combined with clinical information, and further screened lncRNAs related to OS through univariate Cox regression analysis , And used LASSO Cox regression analysis to identify 24 m6A-related lncRNAs that are significantly related to OS; next, a multivariate Cox regression analysis was used to establish a risk model based on 12-m6A-related lncRNAs, and based on the median risk The scores are low-risk group and high-risk group, and Kaplan-Meier is used to analyze the difference in OS between the two groups
.
In addition, drug susceptibility data was used to discover drug candidates targeting these lncRNAs
.
Figure 4.
Flow chart.
Setting the threshold |Pearson R|> 0.
3 & p <0.
001, the author identified 1,149 m6A-related lncRNAs, and the co-expression network is shown in Figure 5
.
Figure 5.
Identifying m6A-related lncRNAs Using univariate Cox regression analysis, 38 m6A-related lncRNAs that are significantly related to OS were screened in the TCGA data set (Figure 6A)
.
LASSO-penalized Cox analysis screened out 24 m6A-related lncRNAs (Figure 6B-C) for subsequent multivariate Cox ratio risk regression analysis, and finally used to construct a risk model to assess the prognostic risk of LUAD patients.
The 12 lncRNAs are shown in Figure 6D Shown
.
Figure 6.
Risk model Figure 7A shows the distribution of risk levels between the low-risk and high-risk groups of the LUAD sample.
The survival status and survival time of the two groups of patients are shown in Figure 7B
.
The results of survival analysis showed that the survival period of the low-risk group was longer than that of the high-risk group (Figure 7D)
.
Figure 7.
The prognostic value of the risk model in the training set is to detect the prognostic performance of the risk model.
The study respectively depicts the risk level distribution, survival status and survival of the test set (Figure 8A-C) and the entire data set (Figure 8E-G) Time pattern, and the expression of m6A-related lncRNAs
.
The Kaplan-Meier survival analysis results of the test set and the entire data set are the same as the training set.
The OS of LUAD patients with higher risk scores is lower than that of patients with lower risk scores (Figure 8D-H)
.
Figure 8.
The prognostic value of the test concentration risk model.
After grouping the entire TCGA data set by gender, age, grade, stage and other clinical indicators, the study found that the OS of the low-risk group was still longer than that of the high-risk group (Figure 9)
.
Figure 9.
Kaplan-Meier survival curve.
Based on the entire gene expression profile, 21 m6A genes, 12 m6A-related lncRNAs, and risk models, the authors performed PCA to analyze the differences between the low-risk group and the high-risk group
.
It is found that in the results obtained based on the risk model, the high and low risk groups have different distributions (Figure 10D)
.
Figure 10.
Principal component analysis Based on the m6A-related lncRNA model obtained from 504 LUAD samples, the author further analyzed the enrichment level, activity and function of several immune cells and pathways in LUAD, and found the immune indicators of the low-risk group and the high-risk group The expression difference was significant (Figure 11A)
.
In addition, through the correlation analysis between the m6A-related lncRNA model and immunotherapy markers, the study found that the high-risk group responded better to immunotherapy (Figure 11C), indicating that the model can be used as a predictor of tumor immune dysfunction and rejection Indicators
.
Next, the author used the R package maftools to analyze the mutation data.
Figure 11D and Figure 11E respectively show the top 20 driver genes with the highest mutation frequency in the high and low risk groups.
.
Then, the TMB score was calculated based on the TGCA somatic mutation data, and the results showed that the classification index has a high correlation with TMB (Figure 11F)
.
The author also used IC50 data in the GDSC database to screen out 78 compounds based on thepRRophetic algorithm as potential therapeutic drugs for LUAD targeting the lncRNA model
.
Figure 11.
Using the model to evaluate immunotherapy response.
The results of univariate and multivariate Cox regression analysis showed that the risk model was independent of other clinical indicators and confirmed its reliability (Figure 12)
.
Figure 12.
Evaluation of the prognostic risk model The authors used a nomogram containing risk levels and clinical risk characteristics to predict the incidence of OS at 1, 2 and 3 years
.
Compared with clinical indicators, the predictive model showed significant predictive ability (Figure 13A)
.
From the correlation diagram in Figure 13B-D, it can be found that the observation rates of 1-year, 2-year, and 3-year OS are consistent with the predicted rates
.
Figure 13.
Construction and evaluation of prognostic nomograms With the development of MeRIP-seq and m6A sequencing technologies, more m6A-modified lncRNAs will be discovered, and their functional significance needs to be explored
.
In addition, research on m6A-related proteins and their inhibitors will also provide new opportunities for early diagnosis, treatment and even prognosis of cancer
.
Therefore, for m6A, there is still a lot of articles to do.
I hope that the articles shared today can bring a bit of inspiration to interested friends
.
Have a nice day! Reference articles: 1.
The role of M6A modification in the regulation of tumor-related lncRNAs (PMID: 33996258) 2.
m6A-related lncRNAs are potential biomarkers for predicting prognoses and immune responses in patients with LUAD (PMID: 33996259) If you have questions about life information, please contact: 18501230653 Dragon Boat Festival Ankang Transcriptome | Methylation | Resequencing | Single Cell | m6A | Multiomics Cytoscape | limma | WGCNA | Water Bear Legend | Linux Electrophoresis | PCR | A Brief History of Sequencing | Karyotype | NIPT | Basic Experimental Genes | 2019-nCoV | Enrichment Analysis | Joint Analysis | Microenvironmental Plague Pursuit | Summary of Ideas | Scholars | Scientific Research | Retracted Papers | PhD Reading | Work