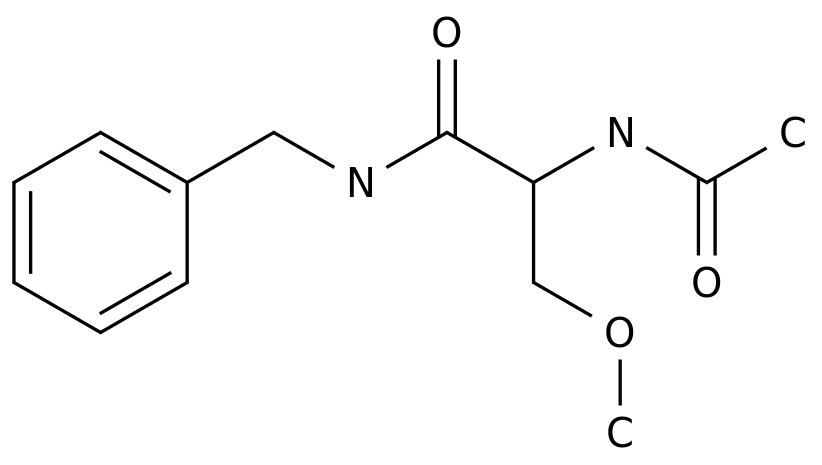
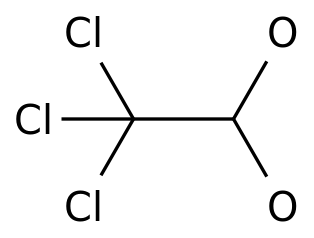
-
Categories
-
Pharmaceutical Intermediates
-
Active Pharmaceutical Ingredients
-
Food Additives
- Industrial Coatings
- Agrochemicals
- Dyes and Pigments
- Surfactant
- Flavors and Fragrances
- Chemical Reagents
- Catalyst and Auxiliary
- Natural Products
- Inorganic Chemistry
-
Organic Chemistry
-
Biochemical Engineering
- Analytical Chemistry
- Cosmetic Ingredient
-
Pharmaceutical Intermediates
Promotion
ECHEMI Mall
Wholesale
Weekly Price
Exhibition
News
-
Trade Service
Study Summary
Stroke is the second leading cause of death worldwide, and previous genome-wide association studies (GWAS) of stroke have been conducted
primarily in populations of European ancestry.
In a cross-ancestry GWAS meta-analysis of 110,182 stroke patients (5 ancestors, 33% non-European) and 1,503,898 control individuals, researchers identified signals associated with stroke and its subtypes on 89 (61 new) independent loci: 60 signals were found in major inverse variance-weighted analyses and 29 signals were found
in secondary meta-regression and multi-feature analyses 。 On the basis of internal cross-ancestry validation and independent follow-up of 89,084 strokes (30% non-European) and 1,013,843 control individuals, they repeated 87% of major stroke risk loci and 60% of minor stroke risk loci (p < 0.
05).
Effect sizes were highly correlated
between different ancestors.
Cross-lineage mapping, microarray mutation analysis, and transcriptome and proteome-wide association analysis further validated the putative disease-causing genes (e.
g.
, SH3PXD2A and FURIN) and their variants (e.
g.
, GRK5 and NOS3).
Using the above three-pronged approach, the researchers provide genetic evidence for the putative drug effect, they propose F11, KLKB1, PROC, GP1BA, LAMC2 and VCAM1 as possible targets for stroke treatment, and researchers are already studying stroke drugs
targeting F11 and PROC.
Polygenic scores that combined cross-bloodline and lineage-specific stroke GWAS with vascular risk factor GWAS (Composite Polygenic Score) effectively predicted ischemic stroke in people of European, East Asian, and African ancestry
.
In 52,600 participants in a clinical trial with cardiometabolic disease, the stroke genetic risk score could accurately predict the risk of ischemic stroke independently of clinical risk
factors.
The findings of this study provide a solid genetic foundation for stroke biology, reveal potential drug targets, and develop tools for predicting stroke genetic risk across ancestors
.
Research background and research methods
Stroke is the second leading cause of death globally, accounting for about 12% of all deaths, especially in low-income countries, where the burden is increasing
.
Stroke is characterized by sudden neurological deficits, mainly caused by cerebral ischemia (the main etiological subtypes are aortic atherosclerotic stroke (LAS), cardiogenic stroke (CES), and small vessel stroke (SVS), and, less commonly
, intracerebral hemorrhage (ICH)).
The higher prevalence of SVS and ICH in Asian and African populations compared with European populations explains the frequency of stroke subtypes in
people of different ancestry.
Most of the gene loci associated with stroke have been identified
in people of European ancestry.
The largest GWAS meta-analysis published to date (67,162 cases and 454,450 control individuals, MEGASTROKE) reported 32 stroke risk loci
.
To identify new genetic associations and broaden the cognitive horizon of stroke pathogenesis and putative drug targets, the researchers first performed cross-bloodline GWAS on 1614080 participants (including 110182 stroke patients), and then repeated previous genome-wide significant locus signals
in separate datasets of 89,084 stroke patients and 1013843 control individuals.
They then identified stroke risk loci by utilizing expression and protein quantity trait loci, cross-ancestor elaborate mapping, and sharing genetic variation with other traits
.
Finally, they used a range of genomics-driven drug discovery methods for stroke prevention and treatment and evaluated the predictive value
of polygenic scores (PGSs) for stroke in the context of population-based studies and clinical trials.
Fig.
2 Effect-size comparison across ancestry groups of lead variants
identified in stroke GWASs and cross-ancestry fine-mapping.
Fig.
3 Genomics-driven drug discovery.
Fig.
4 Risk prediction in a population and trial setting.
Research innovation point one
Eighty-nine (61 new) independent loci
associated with stroke and its subtypes were identified.
F11, KLKB1, PROC, GP1BA, LAMC2 and VCAM1 have been proposed as drug targets for stroke treatment
.
Research innovation point two
Extensive bioinformatics analysis identified genes
that would be prioritized in future functional research.
Such as SH3PXD2A and FURIN
.
Research innovation point three
The results provide genetic evidence for inferring drug effects using three independent methods, two of which (gene enrichment analysis and pQTL-based MR) are consistent
with drug results targeting F11 and KLKB1.
F11 and F11a inhibitors (e.
g.
, abellacimab, BAY 2433334, and BMS-986177) are currently being tested in phase II trials for primary or secondary stroke prevention (NCT04755283, NCT04304508,).
NCT03766581)
。 pQTL-based MR suggests that PROCs are potential drug targets for stroke.
In Phase 1 and Phase 2 trials (3K3A-APC, NCT02222714), recombinant variants of human activated protein C (encoded by PROC) were found to be safe for the treatment of acute ischemic stroke after thrombolysis, mechanical thrombectomy, or both.
and preparing for an upcoming Phase 3 trial
.
3K3A-APC is considered a neuroprotective agent, and there is evidence that it protects white matter bundles and oligodendrocytes from ischemic injury
in mice.
The evidence for GP1BA, VCAM1 and LAMC2 as potential drug targets for stroke is weak, with only one pQTL dataset with evidence
of colocalization.
Under-studied
The study independently validated the vast majority of identified genome-wide significant associations and graded
loci by confidence level based on these findings.
Despite the significant sample size of the follow-up study, with nearly 90,000 stroke patients, this analysis is still not robust, especially for low-frequency variation, ancestry and subtype-specific
associations.
Most follow-up studies in the study came from large biobanks that identified events based on electronic health records without information on suitable stroke subtypes
.
In clinical trial participants with a history of stroke, weak risk predictions may point to selection or indicator event bias and the impact of
secondary prevention treatment.
The main significance of the study
The genomic findings obtained from more than 200,000 stroke patients worldwide have laid an important genetic foundation for future biological studies of stroke pathogenesis, which not only identified potential intervention drug targets, but also provided a powerful tool
for cross-family genetic risk prediction.