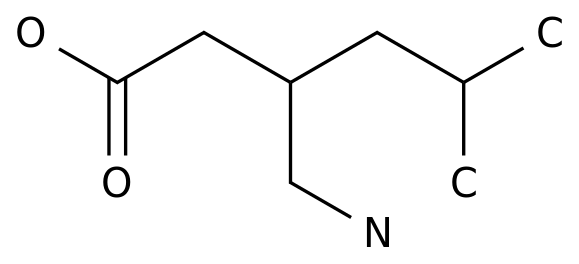
-
Categories
-
Pharmaceutical Intermediates
-
Active Pharmaceutical Ingredients
-
Food Additives
- Industrial Coatings
- Agrochemicals
- Dyes and Pigments
- Surfactant
- Flavors and Fragrances
- Chemical Reagents
- Catalyst and Auxiliary
- Natural Products
- Inorganic Chemistry
-
Organic Chemistry
-
Biochemical Engineering
- Analytical Chemistry
- Cosmetic Ingredient
-
Pharmaceutical Intermediates
Promotion
ECHEMI Mall
Wholesale
Weekly Price
Exhibition
News
-
Trade Service
Why neurodegenerative diseases (Alzheimer's disease, Huntington's disease, Parkinson's disease, Amyotrophic lateral sclerosis, etc.
) appear typical neuronal cell death in specific brain regions has been the focus of the field of neuroscience
.
Small animal models, such as the most commonly used mouse model, are very different from the human brain structure, and most of them cannot simulate the pathological characteristics of typical neuronal cell death in neurodegenerative patients, and drugs that play a role in mouse brain diseases are used in clinical experiments.
Mostly end in failure
.
On March 25, 2022, Professor Li Xiaojiang's team from the Guangdong-Hong Kong-Macao Institute of Central Nervous Regeneration of Jinan University published a review entitled New pathogenic insights from large animal models of neurodegenerative diseases in the journal Protein & Cell (https://link.
springer .
com/article/10.
1007/s13238-022-00912-8), this review summarizes and reviews the advantages and limitations of using large animals such as pigs and monkeys, which are closer to humans, to establish major neurodegenerative disease models
.
Alzheimer's disease (AD), Huntington's disease (HD), Parkinson's disease (PD), Amyotrophic Lateral Sclerosis (ALS), etc.
are all caused by protein It is a neurodegenerative disease that seriously threatens human health in today's society
.
These diseases produce progressive neuronal cell death with aging
.
Due to the lack of suitable animal models for drug screening, there is currently no effective treatment
.
The establishment of animal models that accurately mimic disease pathology and phenotype is crucial for the development of effective means of treating diseases
.
The authors first compared the species differences between mice, pigs, monkeys and humans in terms of brain structure, development, number of nerve cells, etc.
, and summarized the neurodegenerative disease large animals (pig) established by CRISPR/Cas9 gene editing technology in recent years.
and monkey) models and the important results based on these models, taking HD, PD, ALS large animal models as examples, showing that large animal models can better simulate the important pathological characteristics of neurodegeneration in the patient's brain
.
For example, Huntington's disease (HD) is a neurodegenerative disorder caused by mutations in a single gene (HTT) containing abnormally long glutamine repeats
.
However, the mutant HTT knock-in mouse model did not exhibit the typical neuronal cell death characteristics in the patient's brain
.
The HTT gene knock-in pig model not only mimics the typical pathological features of specific neuronal death in the brain of HD patients, but also exhibits abnormal motor behaviors similar to HD
.
More importantly, these pathological features and abnormal behaviors can be passed on to offspring, providing a new and important animal model for the treatment of HD
.
Parkinson's disease (PD) has long been considered to be related to the damage of the organelle (mitochondria) that supplies energy in cells, and PINK1, as a mitochondrial membrane protein, is recruited to damaged mitochondria to activate Parkin and ubiquitinase to form a complex , so that damaged mitochondria can be promptly removed by lysosomes, thereby protecting nerve cells
.
This classic theory is mainly established based on a large number of in vitro experiments
.
However, PINK1 is difficult to detect in mouse brains, and PINK1-knockout mice cannot confirm the above theory nor mimic the pathological features of neuronal cell death in patients' brains
.
The PINK1 knockdown non-human primate model established by CRISPR/Cas9 technology can simulate the pathological characteristics of neuronal cell death in Parkinson's patients, and reveal that PINK1 kinase is abundantly expressed in primate brain, phosphorylating a series of important neural functions protein to maintain the survival of nerve cells (experts commented on Protein & CellSubverting the classic theory, Li Xiaojiang's team revealed a new mechanism of hereditary Parkinson's disease) (Figure 1)
.
Therefore, studies in large animal models have shown that PINK1 has an important kinase function other than mitochondrial function, which provides a new research direction for the study of multiple functions of PINK1 and PD
.
Figure 1: The classical theory that PINK1 is involved in mitophagy is mainly based on the results of a large number of in vitro cultured cell experiments, while non-human primate experiments prove that PINK1 is mainly a kinase to maintain neuronal survival
.
TDP-43 is a pathogenic protein of neurodegenerative diseases.
It is normally distributed in the nucleus of nerve cells and participates in important processes such as gene transcription inhibition and RNA precursor splicing
.
However, in neuropathological conditions such as amyotrophic lateral sclerosis (ALS), TDP-43 in the nucleus is transferred to the cytoplasm, causing neurotoxicity
.
However, in the mouse model, the vast majority of TDP-43 still accumulates in the nucleus, which cannot fully simulate the pathological characteristics of TDP-43 accumulation in the cytoplasm in the brain of patients
.
However, the hydrolase caspase-4, which is specifically expressed in higher animals such as humans and monkeys, can cleave TDP-43, so that the TDP-43 fragment product lacking the nuclear localization signal is transferred to the cytoplasm, which is similar to the cytoplasmic TDP-43 in patients.
The pathological features of aggregation are distinct from the accumulation of TDP-43 in the nucleus observed in mouse models
.
In addition, TDP-43 in the cytoplasm can specifically bind to the (TG)n repeat in the 3'UTR of the primate autophagy-regulating multifunctional protein p62/SQSTM1 gene, promoting the instability of the monkey SQSTM1 gene and affecting its function ( Figure 2)
.
However, the 3'UTR region of p62/SQSTM1 in mice lacks longer (TG)n repeats and is not bound by TDP-43, once again demonstrating the importance of using non-human primate models to study neurodegenerative diseases
.
Figure 2: Primate-specific hydrolase caspase-4 cleaves TDP-43, translocating the TDP-43 fragment product lacking the nuclear localization signal to the cytoplasm and binding to p62/SQSTM1 mRNA, resulting in cytoplasmic toxicity
.
The authors pointed out that small animal models have made important contributions to pathogenic mechanism research and preclinical treatment research, but the expression and function of some important pathogenic genes are species-specific, so small animal models cannot simulate important pathological characteristics of patients The causative gene can be considered to be studied using large animal models
.
However, the use of large animal models also has significant limitations
.
Large animals have a long reproductive cycle and are expensive, and gene modification technology is still not as complete as the establishment of small animal models.
In particular, off-target effects and chimeric effects caused by gene editing technology will also affect the batch establishment of large animal disease models with consistent genetic characteristics
.
Therefore, the authors also discuss how to optimize the establishment of models for these limitations, for example, genetic modification can be directly performed in the adult large animal brain to observe pathological changes, conduct molecular mechanism studies, and find key pathological and therapeutic targets
.
With the continuous progress and development of new technologies such as single-cell analysis technology, high-throughput data analysis, and microscopic imaging technology, combined with the continuous optimization of gene editing technology, there will be more large animals that can better simulate neurodegenerative diseases in the future.
Models have emerged, providing important animal models for the treatment of neurodegenerative diseases
.
Researcher Yin Peng, Professor Li Shihua, Professor Li Xiaojiang and Researcher Yang Weili of Jinan University Guangdong-Hong Kong-Macao Institute of Central Nervous Regeneration are the co-authors of this review
.
Original link: https://doi.
org/10.
1007/s13238-022-00912-8 Publisher: 11th reprint notice [Non-original article] The copyright of this article belongs to the author of the article, and personal sharing is welcome.
Reprinting is prohibited without permission.
The author has all legal rights, and violators will be prosecuted
.
) appear typical neuronal cell death in specific brain regions has been the focus of the field of neuroscience
.
Small animal models, such as the most commonly used mouse model, are very different from the human brain structure, and most of them cannot simulate the pathological characteristics of typical neuronal cell death in neurodegenerative patients, and drugs that play a role in mouse brain diseases are used in clinical experiments.
Mostly end in failure
.
On March 25, 2022, Professor Li Xiaojiang's team from the Guangdong-Hong Kong-Macao Institute of Central Nervous Regeneration of Jinan University published a review entitled New pathogenic insights from large animal models of neurodegenerative diseases in the journal Protein & Cell (https://link.
springer .
com/article/10.
1007/s13238-022-00912-8), this review summarizes and reviews the advantages and limitations of using large animals such as pigs and monkeys, which are closer to humans, to establish major neurodegenerative disease models
.
Alzheimer's disease (AD), Huntington's disease (HD), Parkinson's disease (PD), Amyotrophic Lateral Sclerosis (ALS), etc.
are all caused by protein It is a neurodegenerative disease that seriously threatens human health in today's society
.
These diseases produce progressive neuronal cell death with aging
.
Due to the lack of suitable animal models for drug screening, there is currently no effective treatment
.
The establishment of animal models that accurately mimic disease pathology and phenotype is crucial for the development of effective means of treating diseases
.
The authors first compared the species differences between mice, pigs, monkeys and humans in terms of brain structure, development, number of nerve cells, etc.
, and summarized the neurodegenerative disease large animals (pig) established by CRISPR/Cas9 gene editing technology in recent years.
and monkey) models and the important results based on these models, taking HD, PD, ALS large animal models as examples, showing that large animal models can better simulate the important pathological characteristics of neurodegeneration in the patient's brain
.
For example, Huntington's disease (HD) is a neurodegenerative disorder caused by mutations in a single gene (HTT) containing abnormally long glutamine repeats
.
However, the mutant HTT knock-in mouse model did not exhibit the typical neuronal cell death characteristics in the patient's brain
.
The HTT gene knock-in pig model not only mimics the typical pathological features of specific neuronal death in the brain of HD patients, but also exhibits abnormal motor behaviors similar to HD
.
More importantly, these pathological features and abnormal behaviors can be passed on to offspring, providing a new and important animal model for the treatment of HD
.
Parkinson's disease (PD) has long been considered to be related to the damage of the organelle (mitochondria) that supplies energy in cells, and PINK1, as a mitochondrial membrane protein, is recruited to damaged mitochondria to activate Parkin and ubiquitinase to form a complex , so that damaged mitochondria can be promptly removed by lysosomes, thereby protecting nerve cells
.
This classic theory is mainly established based on a large number of in vitro experiments
.
However, PINK1 is difficult to detect in mouse brains, and PINK1-knockout mice cannot confirm the above theory nor mimic the pathological features of neuronal cell death in patients' brains
.
The PINK1 knockdown non-human primate model established by CRISPR/Cas9 technology can simulate the pathological characteristics of neuronal cell death in Parkinson's patients, and reveal that PINK1 kinase is abundantly expressed in primate brain, phosphorylating a series of important neural functions protein to maintain the survival of nerve cells (experts commented on Protein & CellSubverting the classic theory, Li Xiaojiang's team revealed a new mechanism of hereditary Parkinson's disease) (Figure 1)
.
Therefore, studies in large animal models have shown that PINK1 has an important kinase function other than mitochondrial function, which provides a new research direction for the study of multiple functions of PINK1 and PD
.
Figure 1: The classical theory that PINK1 is involved in mitophagy is mainly based on the results of a large number of in vitro cultured cell experiments, while non-human primate experiments prove that PINK1 is mainly a kinase to maintain neuronal survival
.
TDP-43 is a pathogenic protein of neurodegenerative diseases.
It is normally distributed in the nucleus of nerve cells and participates in important processes such as gene transcription inhibition and RNA precursor splicing
.
However, in neuropathological conditions such as amyotrophic lateral sclerosis (ALS), TDP-43 in the nucleus is transferred to the cytoplasm, causing neurotoxicity
.
However, in the mouse model, the vast majority of TDP-43 still accumulates in the nucleus, which cannot fully simulate the pathological characteristics of TDP-43 accumulation in the cytoplasm in the brain of patients
.
However, the hydrolase caspase-4, which is specifically expressed in higher animals such as humans and monkeys, can cleave TDP-43, so that the TDP-43 fragment product lacking the nuclear localization signal is transferred to the cytoplasm, which is similar to the cytoplasmic TDP-43 in patients.
The pathological features of aggregation are distinct from the accumulation of TDP-43 in the nucleus observed in mouse models
.
In addition, TDP-43 in the cytoplasm can specifically bind to the (TG)n repeat in the 3'UTR of the primate autophagy-regulating multifunctional protein p62/SQSTM1 gene, promoting the instability of the monkey SQSTM1 gene and affecting its function ( Figure 2)
.
However, the 3'UTR region of p62/SQSTM1 in mice lacks longer (TG)n repeats and is not bound by TDP-43, once again demonstrating the importance of using non-human primate models to study neurodegenerative diseases
.
Figure 2: Primate-specific hydrolase caspase-4 cleaves TDP-43, translocating the TDP-43 fragment product lacking the nuclear localization signal to the cytoplasm and binding to p62/SQSTM1 mRNA, resulting in cytoplasmic toxicity
.
The authors pointed out that small animal models have made important contributions to pathogenic mechanism research and preclinical treatment research, but the expression and function of some important pathogenic genes are species-specific, so small animal models cannot simulate important pathological characteristics of patients The causative gene can be considered to be studied using large animal models
.
However, the use of large animal models also has significant limitations
.
Large animals have a long reproductive cycle and are expensive, and gene modification technology is still not as complete as the establishment of small animal models.
In particular, off-target effects and chimeric effects caused by gene editing technology will also affect the batch establishment of large animal disease models with consistent genetic characteristics
.
Therefore, the authors also discuss how to optimize the establishment of models for these limitations, for example, genetic modification can be directly performed in the adult large animal brain to observe pathological changes, conduct molecular mechanism studies, and find key pathological and therapeutic targets
.
With the continuous progress and development of new technologies such as single-cell analysis technology, high-throughput data analysis, and microscopic imaging technology, combined with the continuous optimization of gene editing technology, there will be more large animals that can better simulate neurodegenerative diseases in the future.
Models have emerged, providing important animal models for the treatment of neurodegenerative diseases
.
Researcher Yin Peng, Professor Li Shihua, Professor Li Xiaojiang and Researcher Yang Weili of Jinan University Guangdong-Hong Kong-Macao Institute of Central Nervous Regeneration are the co-authors of this review
.
Original link: https://doi.
org/10.
1007/s13238-022-00912-8 Publisher: 11th reprint notice [Non-original article] The copyright of this article belongs to the author of the article, and personal sharing is welcome.
Reprinting is prohibited without permission.
The author has all legal rights, and violators will be prosecuted
.