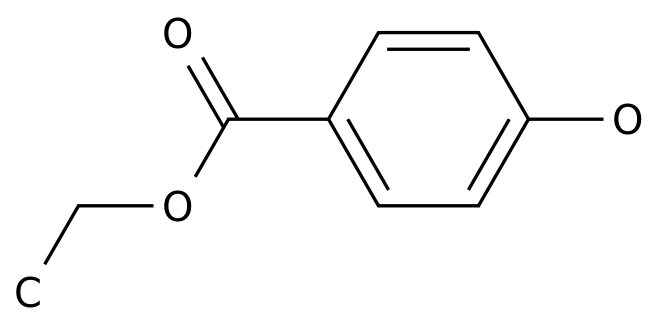
-
Categories
-
Pharmaceutical Intermediates
-
Active Pharmaceutical Ingredients
-
Food Additives
- Industrial Coatings
- Agrochemicals
- Dyes and Pigments
- Surfactant
- Flavors and Fragrances
- Chemical Reagents
- Catalyst and Auxiliary
- Natural Products
- Inorganic Chemistry
-
Organic Chemistry
-
Biochemical Engineering
- Analytical Chemistry
-
Cosmetic Ingredient
- Water Treatment Chemical
-
Pharmaceutical Intermediates
Promotion
ECHEMI Mall
Wholesale
Weekly Price
Exhibition
News
-
Trade Service
How to use Crystal Miner analysis software to analyze Drop-off digital PCR results
Only two channels of Crystal Digital PCR can be used to perform absolute quantification of mutant alleles (MAF) with high sensitivity
The first channel: The Drop-Off probe is only complementary to the wild-type sequence, and the mutant sequence is skipped
The second channel: the reference probe hybridizes to the adjacent site of the mutation site and complements the mutation and wild-type alleles (the green channel on the horizontal axis in Figure 1)
When a wild-type allele is present, both the drop-off and the reference probe will hybridize with the template, and a double positive droplet will appear
When there is a mutant allele, even a single nucleotide mutation is enough to destroy the instability of the drop-off probe hybridization, so only the reference probe can hybridize to the template, thereby generating a single positive spot
For more information on Drop-off design and analysis, please refer to our manual "Drop-off Crystal Digital PCR Detection of NRAS, KRAS and EGFR Mutations", website, https:// /
A.
B.
Use Crystal Miner analysis software to quantitatively analyze Drop-off results step by step
The wild-type concentration Cwt (double positive droplets) can be calculated directly by the ratio of Pwt (regardless of whether the third channel is positive or negative), and the concentration of drop-off mutation Cmut (mutant allele) can be calculated by Pmut
To perform quantitative calculations using Crystal Miner analysis software, you need to follow the steps below:
1.
Follow the steps, ANALYZE DATA> Plots&Populations> 2D dot plot
2.
Steps: ANALYZE DATA> Plots&Populations> Population Editor
3.
hint:
The LOD value relative to the drop-off mutant increases exponentially with the expected wild-type concentration (at the 95% confidence level, the LOD increases from 0.
Gene-π Digital PCR School:
More digital PCR technologies can be viewed at Gene-π Digital PCR School (Digital PCR School Quick Entry).
Contact us: