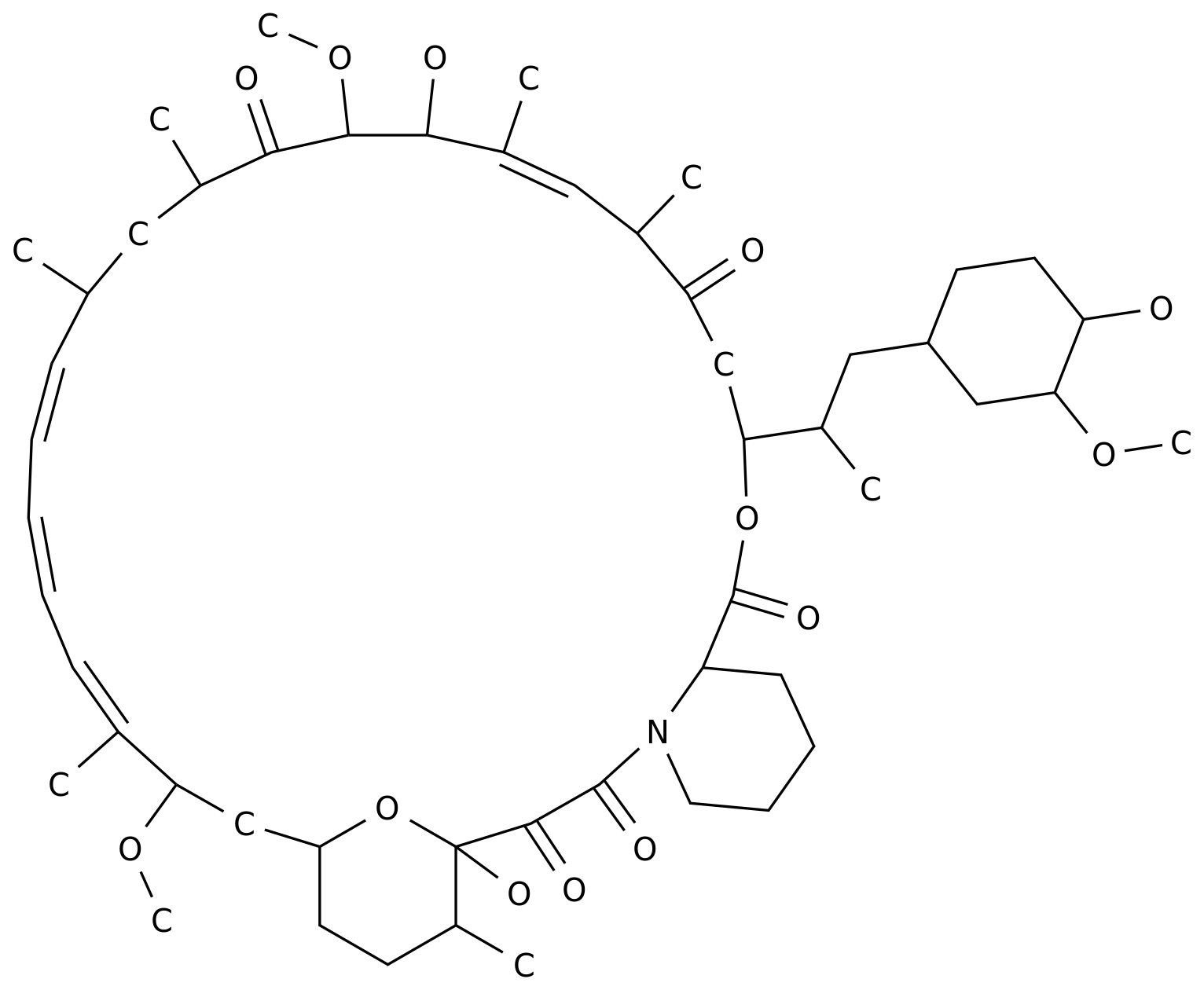
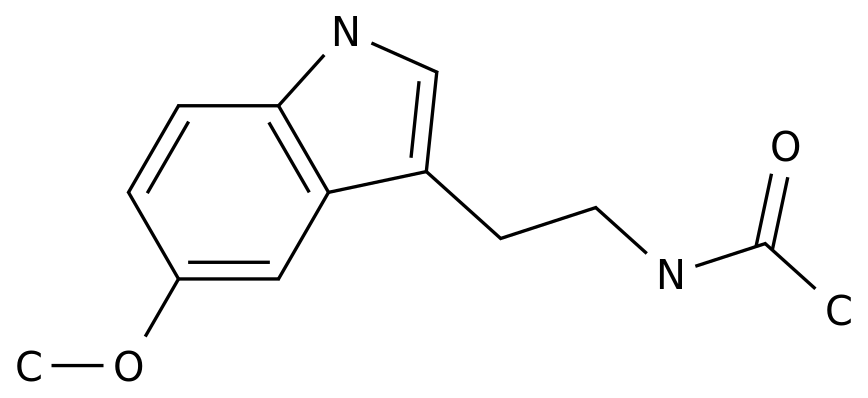
-
Categories
-
Pharmaceutical Intermediates
-
Active Pharmaceutical Ingredients
-
Food Additives
- Industrial Coatings
- Agrochemicals
- Dyes and Pigments
- Surfactant
- Flavors and Fragrances
- Chemical Reagents
- Catalyst and Auxiliary
- Natural Products
- Inorganic Chemistry
-
Organic Chemistry
-
Biochemical Engineering
- Analytical Chemistry
- Cosmetic Ingredient
-
Pharmaceutical Intermediates
Promotion
ECHEMI Mall
Wholesale
Weekly Price
Exhibition
News
-
Trade Service
OBJECTIVE : Genome-wide association studies (GWAS) have identified more than 100 risk loci for systemic lupus erythematosus (SLE) , but disease genes at most loci remain unclear, hindering translation of these gene discoveri.
The study aimed to prioritize genes for the 110 SLE loci identified in the latest East Asian GWAS meta - analysi.
The study aimed to prioritize genes for the 110 SLE loci identified in the latest East Asian GWAS meta - analysi.
Methods : We established gene expression prediction models in blood B cells, CD4+ and CD8+ T cells, monocytes, natural killer cells, and peripheral blood cells of 105 Japanes.
Transcriptome-wide association studies (TWAS) were performed using data from a recent genome-wide association meta-analysis of 208,370 East Asians , and candidate genes were searched using TWAS and three data-driven computational approache.
Transcriptome-wide association studies (TWAS) were performed using data from a recent genome-wide association meta-analysis of 208,370 East Asians , and candidate genes were searched using TWAS and three data-driven computational approache.
Results : TWAS identified 171 SLE genes ( p<0 × 10 -5 ); 114 (67%) were significant only in a single cell type; 127 (73%) were located in the SLE GWAS locu.
TWAS identified a strong association between CD83 and SLE ( p< 7 × 10-8 .
A meta - analysis of genomic associations in existing 208,370 East Asians and an additional 1,498 SLE cases and 3,330 controls found rs72836542 around CD83 ( OR =11 , p=5 × 10-9
) with a new single-gene variant associati.
For the 110 SLE loci , the researchers identified 276 candidate genes, including 104 genes at the recently discovered new SLE loc.
It was demonstrated in vitro that the putative causal variant rs61759532 exhibited allele-specific regulation of ACAP1 and that the presence of the SLE risk allele reduced ACAP1 expressio.
TWAS identified a strong association between CD83 and SLE ( p<7 × 10 -8 -8 .
A meta-analysis of genomic associations in existing 208,370 East Asians and an additional 1,498 SLE cases and 3,330 controls found rs72836542 around CD83 CD83 ( OR =11 ,
There was a new single-gene variant association at p=5 × 10 -9 -9 )
It was demonstrated in vitro that the putative causal variant rs61759532 exhibits allele-specific regulation of ACAP1 ACAP1 and that the presence of the SLE risk allele reduces ACAP1 ACAP1 expressio.
Conclusions : Cell-level transcriptome-wide association studies in six immune cells complement the SLE gene discovery and guide the identification of novel genetic associatio.
These gene findings provide biological insights into the genetic associations of SL.
These gene findings provide biological insights into the genetic associations of SL.
Source: Yin X, Kim K, Suetsugu H , et a.
Biological insights into systemic lupus erythematosus through an immune cell-specific transcriptome-wide association stu.
Source: Yin X, Kim K, Suetsugu H , et a.
Biological insights into systemic lupus erythematosus through an immune cell-specific transcriptome-wide association stu.
Annals of the Rheumatic DiseasesPublished Online First: 24 May 202 doi: 11136/annrheumdis -2022-222345 , et al Annals of the Rheumatic Diseasesleave a message here